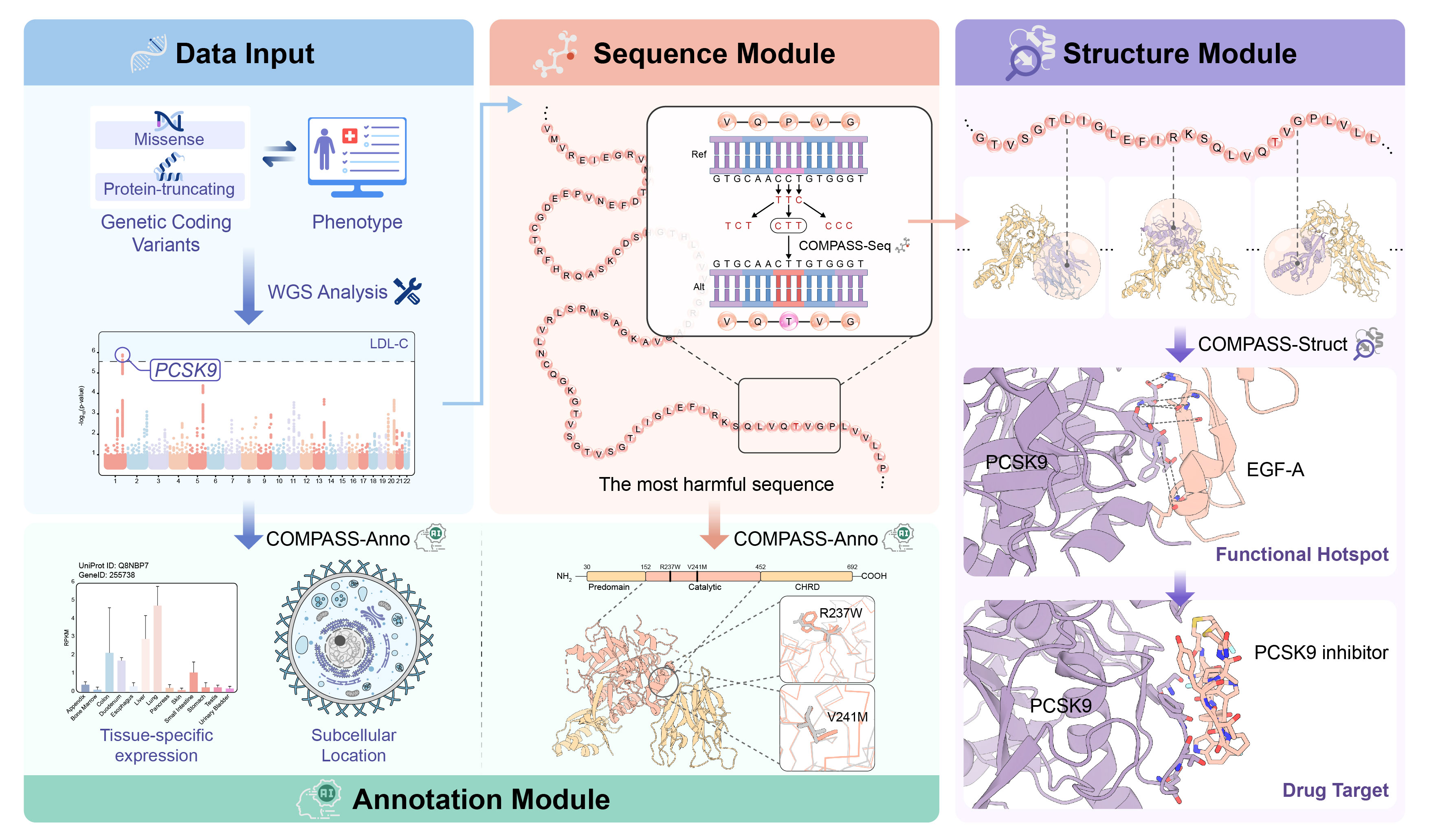
Overview
COMPASS is a cutting-edge computational pipeline designed to systematically evaluate the functional and structural consequences of disease-associated coding variants. By integrating genetic association analysis with AI-predicted protein structures, this innovative approach bridges the gap between genetic variation and phenotypic outcomes, offering new insights into disease mechanisms and potential therapeutic targets.
Our Mission
Our goal is to enhance the understanding of how genetic variants contribute to complex diseases and traits, ultimately accelerating the discovery of drug targets. COMPASS provides a scalable and comprehensive framework to uncover actionable genetic insights for precision medicine.
What We Do
COMPASS employs a multi-step methodology to analyze coding variants:
Sequence-based Selection: Identifies disease-associated coding variants based on statistical significance from whole-genome sequencing studies. These variants are then mapped to transcript-based amino acid sequences and selects reprensentive animo acid sequence.
Structure-based patch scanning: Uses a structure-based patch scanning approach to pinpoint disease-relevant protein subregions.
Therapeutic Insights: By integrating structural and functional analyses of disease-associated variants, COMPASS pinpoints regions of potential therapeutic relevance.
Key Applications
COMPASS has been applied to the analysis of rare coding variants associated with pan-cancer phenotypes in the UK Biobank 500K whole-genome sequencing data, revealing structural hotspots that underlie disease mechanisms and therapeutic potential. Among the identified genes, JAK2 exhibited a strong association with polycythemia vera (PV) (P = 3.81E-237). By integrating sequence-based selection and structure-based patch scanning, COMPASS precisely localized disease-relevant regions within the JH1 kinase and JH2 pseudokinase domains, including the well-characterized V617F mutation site and the ATP-binding pocket. These structural hotspots closely correspond to the binding sites of approved JAK2 inhibitors such as ruxolitinib and fedratinib, as well as the investigational JH2-targeting compound flonoltinib maleate. Beyond PV, COMPASS identified shared JAK2 structural hotspots across leukemia and non-Hodgkin’s lymphoma, indicating cross-disease convergence in structural and functional disrupts. These findings validate the robustness of COMPASS and demonstrate its ability to pinpoint functional hotspots and druggable regions, providing a structural basis for therapeutic discovery and drug repurposing.
Why It Matters
Improved Drug Discovery: By linking disease-associated genetic variants to structural and functional protein changes, COMPASS supports the identification of high-confidence drug targets, which are associated with higher clinical trial success rates.
Precision Medicine: Provides a deeper understanding of disease mechanisms at the molecular level, paving the way for personalized therapeutic strategies.
Scalable and Versatile: The pipeline is designed to be adaptable across various diseases and traits, making it a powerful tool for genomic research.